About
Me, myself and I
If you've ended up here you must be quite nosy. So here's your todays' treat. My name's Daniel and I'm a plant molecular geneticist. I studied plain old biology however I rather quickly shifted my main emphasis towards plants. I can get obsessed with details which is why I guess molecular biology was very tempting for me. After finishing my studies I went and did a PhD in wine consumption in the South-West of Germany. Jokes aside I did my PhD in an applied field of plant breeding. To be precise in breeding fungal resistant grapevine cultivars. I studied the interaction of fungal resistant grapevine cultivars and the grapevine powdery mildew pathogen.
I enjoy the freedom of research. Having the opportunity to look into things nobody has ever done in such detail before makes me feel humble and grateful. The great unkown of most of the things in biology has therefore a special attraction for me.
But that's only one of the things that I enjoy doing. The other passion that I have is building things and taking things apart to learn how they work. I remember when I was little playing with the tools of my grandpa who was a heating fitter. Soldering together scrap pipes (and wasting his supplies), using other scraps to build little racing cars is what was keeping me busy all day long and are the things sticking to my mind when I think back. Over the years I forgot about this and it took quite some time to rediscover this passion. But nowadays I love playing around with 3D printers and the endless possiblilities the arduino micro cosmos has in store.
I always had a certain affinity towards computers. Staying up late, staring into the screen, playing online with friends was a great pleasure. It's no surprise that today the computer screen plays a central role in my life. I went down the rabbit hole of computational biology at the end of my PostDoc. It's impressive how busy you can be with compiling the dependencies of published pipelines... Anyhow, I love playing around with the possibilities of available open-source tools for the analysis of genetic data (be it DNA, RNA or proteins) or image data for automatic phenotyping.
I guess this website should house the things that I like to do. What good is it to learn somthing and then don't share the process and the final results of it?! You won't get a medal for taking your knowledge to the grave eventually. So everything that I share is open for you to play with, adapt it to your needs and make it available to others in the same way. Everybody is thankful about some help sometimes (I'm no exception there!). If you find any of the things on this website helpful let me know. If you find any thing on this website to be crap let me know, too. We will then figure out together how to make crap shine golden ;)
Cheers,
Daniel
Scientific publications
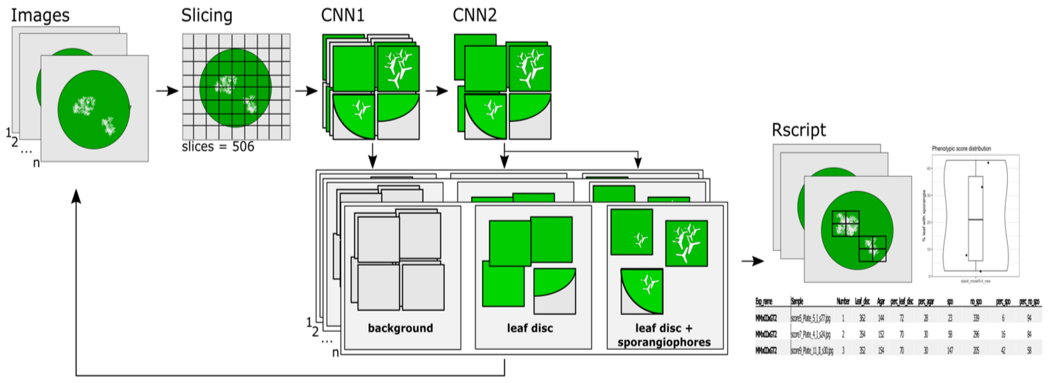
High-throughput phenotyping of leaf discs infected with grapevine
downy mildew using shallow convolutional neural networks
Zendler D.; Malagol N.; Schwandner A.; Töpfer R.; Hausmann L.; Zyprian E.
Agronomy, 2021, 11(9), 1768; doi:10.3390/agronomy11091768
GitHub repository: Daniel-Ze/Leaf-disc-scoring
Preprint: bioRxiv 2021, 08, doi:10.1101/2021.08.19.456931
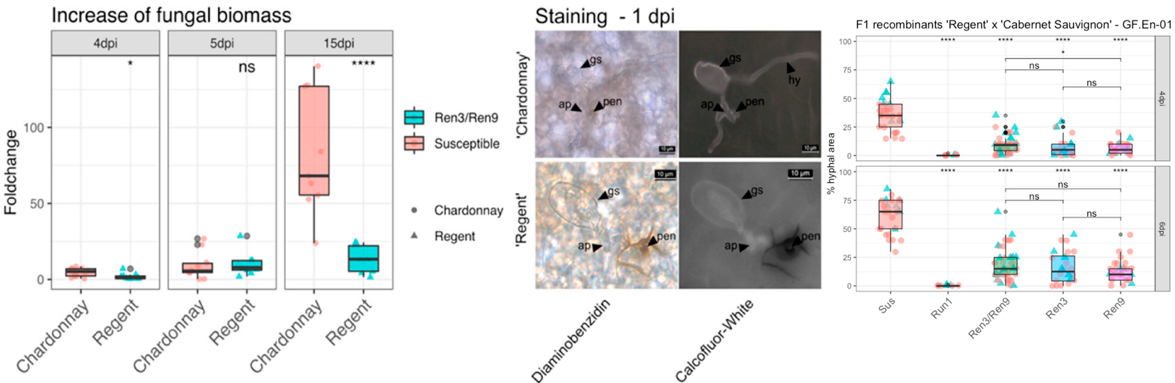
Confirmation and Fine Mapping of the Resistance Locus Ren9 from the Grapevine Cultivar 'Regent'
(Editor's choice article)
Zendler, D.; Töpfer, R.; Zyprian, E.
Plants, 2021, 10(1), 24; doi:10.3390/plants10010024
Preprint: bioRxiv 2020, 11, doi:10.1101/2020.11.27.400770
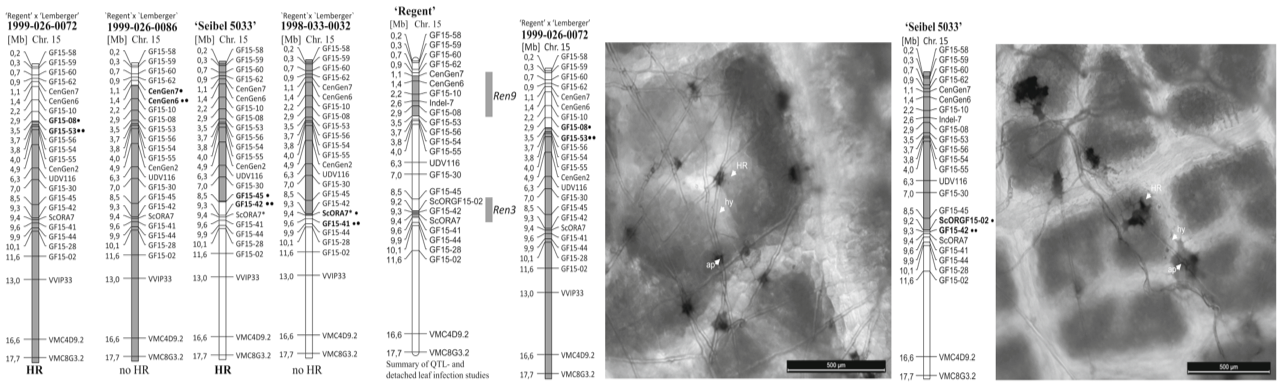
Fine mapping of Ren3 reveals two loci mediating hypersensitive response against Erysiphe necator in grapevine.
Zendler, D.; Schneider, P.; Töpfer, R.; Zyprian, E.
Euphytica, 2017, 213, 68; doi:10.1007/s10681-017-1857-9
RNA Processing Factor 7 and Polynucleotide Phosphorylase Are Necessary for Processing and Stability of nad2 mRNA in Arabidopsis Mitochondria.
Stoll, B.; Zendler, D.; Binder, S.
RNA Biology, 2014, 11, P.968-976; doi:10.4161/rna.29781
