Run everything in parallel!
18.06.2021
GNU parallel
So recently I was playing with Raspberry Pi 4s. Beginning of the year I bought one with 4gb of RAM and it is running Pi-Hole and a MQTT server in my network. Pi-hole is some amazing thing. I really don't understand why I set this up only now... Anyways, during that time I saw that the model 4 of the Pis can have up to 8gb of RAM which according to people on amazon reviews is absolutely bonkers. But is it really?! So I decided to grab a bit of gold from my vault to buy three of these little computers as I wanted to try out some cluster computation software like SLURM or open MPI. And that's were it hit me. I was using a software called GNU parallel [1] for quite some time in my analyses with tools which don't have the built in ability of running things in parallel (like Augustus the gene prediction software). This neat piece of software allows me not going through the trouble of gaining knowledge in multithreading and multiprocessing!! Looking in the manual of GNU parallel I realized that this software allows SSH access to remote machines to run programs on these remote machines (in parallel of course!).
Running GNU parallel
Running this requires you to have set up passwordless SSH access to your remote server / nodes. To get this going follow this tutorial: Setup passwordless SSH
Another prerequisite for this to work is the availability of GNU parallel on all the nodes of your soon to be cluster. Chose what suits you the most:
Ubuntu, Raspbian: sudo apt install parallel MacOS: brew install parallel conda install parallel mamba install parallel
Further you must ensure that the program that you want to run is available on all the nodes as well. In our case this will be echo which is a standard program available on all Unix systems. The easiest way of remote parallel computation is shown below:
parallel -S 4/daniel@1.1.1.100 echo "Number {}: Running on \`hostname\`" ::: 1 2 3 4 Number 1: Running on server Number 2: Running on server Number 4: Running on server Number 3: Running on server
Remember that this is parallel executed therefore the sequence of numbers most probably won’t be printed in the right order. So with this we executed a command in parallel on a remote machine! Wow! Now what’s exactly happening here?
Remote server:
-S CPUs/Username@server
Place holder for input:
{} = Input variable
Actual input:
::: 1 2 3 4
We are running the echo command on the remote machine in parallel. We defined that parallel would use 4 CPU cores on this machine and we gave a sequence of numbers (1 2 3 4) which is the input ({}) in the echo command.
So this was an easy remote and parallel execution of what would usually have been a serial for loop after ssh login. To make this whole thing more cluster-like parallel has a flag –sshloginfile in which we can store the cluster nodes:
cat ~/nodelist 4/ daniel@1.1.1.100 4/ daniel@1.1.1.101 4/ daniel@1.1.1.102 8/ daniel@1.1.1.199
This would represent my three raspberry Pis and my little home-server for which I have setup passwordless SSH login. Running the above command using the nodelist:
parallel --sshloginfile ~/nodelist echo "Number {}: Running on \`hostname\`" ::: 1 2 3 4 Number 1: Running on server Number 2: Running on pi8gb-cluster-2 Number 3: Running on pi8gb-cluster-3 Number 4: Running on pi8gb-cluster-1
By chance the sequence of numbers was printed in the right order this time. However, the difference to the above command can be seen now with the printed hostnames. One echo comes from the home server and the other echos come from the three little Raspberry Pis! Unbelievable! We just echoed a sequence of numbers in a cluster! This little echo command is the perfect test if all your nodes can be reached and everything’s setup properly.
Practical use scenario
As you have proven that you can run programs remotely in parallel on your cluster nodes we can step up the game a notch. How about aligning protein sequences against a genome for which you don’t have a proper gene annotation yet? Let’s do it!
First we make sure that we have the program installed which will do the alignment of the protein data against the genome:
sudo apt install exonerate
Repeat this on all the nodes in your cluster and bam! we are all set. Assuming we have a genome consisting of chromosomes in a multi-fasta file and another multi-fasta file with thousands of protein sequences at hand we can start our little test. To leverage the power of parallelization we have to split one of the two multi-fasta files into smaller chunks. I will go with the protein file in my example:
mkdir /home/daniel/sequences/protein/test/tmp python split_chunk.py protein.fa 1000 test /home/daniel/sequences/protein/test/
I wrote this python script to read in a multi-fasta (protein.fa) file and split into n chunks (1000) which are named test and they are stored in a folder called tmp. Now that we have split the multi-fasta protein file into 1000 chunks we have to do one last thing. As the genome file will be a constant in our analysis we have to make sure that the file is available on all the nodes. We can do this manually by using scp:
scp genome.fa daniel@1.1.1.1XX:/home/daniel
Repeat this for all the nodes and we’re finally all set. Let’s do it! Computation on all the nodes!1!11!
cd /home/daniel/sequences/test/temp /home/daniel/sequences/test/temp $ ls *.fa | parallel --progress \ --sshloginfile ~/nodelist \ --trc {}.exonerate.gff \ --retries 4 \ --resume-failed \ --joblog parallel.log \ exonerate --model protein2genome {} ./genome.fa \ --showtargetgff yes \ --bestn 5 \ --percent 90 ">>"{}.exonerate.gff
Now that’s quite a load of arguments you might say. Don’t worry I’ll explain them in detail. Be aware that the above is a one line command!
GNU parallel commands explained:
--progress = Show the progress on how many jobs are completed by each node
--sshloginfile = This is were we indicate our node list with CPU/ user@ip
--trc = This one is mighty. It stands for transfer, return and cleanup.
parallel is transfering the piped in protein file to the nodes.
After the nodes are finished with their job parallel returns
the file and removes it from the host (cleanup). in our case
the {}.exonerate.gff is the output of exonerate that we specify
later.
--retries = Assuming something goes sideways parallel has you covered!
--joblog Together with the --resume-failed flag and by turning on the --joblog
--resume-failed parallel can keep track of what's happening. When a job fails,
gets killed it is noted in the log file. The failed jobs are
automatically resumed at the end of the run.
Exonerate commands explained:
--model = protein2genome is quite self explaining ;) The {} is again the
input placeholder described above. In this case it represents
the piped input protein sequence file. Together with the
protein file the genome.fa file is indicated which was transferred
to all the nodes and lies in the home folder of the user daniel.
--showquerygff = This will output a gff version 2 annotation of the aligned
protein sequence.
--bestn = Output only the best 5 matches
--percent = Only output matches with 90% sequence similarity
There are a few things that are crucial for the above to work. (1) The genome file needs to be located in the home folder of the user with which we SSH in remotely. By SSH login we end up in the users home folder and parallel is not doing anything different here. (2) make sure that the –trc {}.exonerate.gff and the >> {}.exonerate.gff at the end match 100%. Parallel is returning exactly what you define it to return! If you got all that right you might see something like this depending on your cluster setup at home:
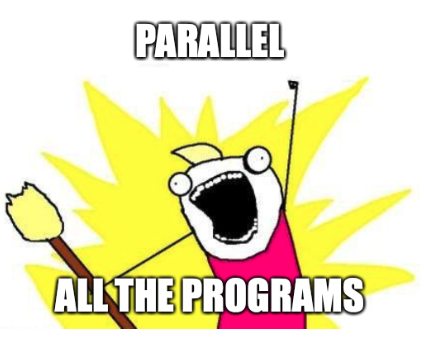
ls *.fa| parallel --gnu --progress --sshloginfile ~/nodelist --trc {}.exonerate.gff --retries 4 --resume-failed --joblog parallel_cluster_1.log exonerate --model protein2genome {} ./genome.fa --showtargetgff yes --bestn 5 --percent 90 ">>"{}.exonerate.gff Computers / CPU cores / Max jobs to run 1: daniel@192.168.178.100 / 3 / 3 2: daniel@192.168.178.101 / 3 / 3 3: daniel@192.168.178.102 / 3 / 3 4: daniel@192.168.178.199 / 8 / 8 Computer:jobs running/jobs completed/%of started jobs/Average seconds to complete daniel@1.1.1.100:3/5/7%/4327.4s daniel@1.1.1.101:3/5/7%/4327.4s daniel@1.1.1.102:3/4/6%/5409.2s daniel@1.1.1.199:8/70/77%/309.1s

The Raspberry Pi cluster on top of my server.
As you can see all nodes are doing their job (more or less fast ... :D). The poor Raspberry Pis are no match for the AMD Rayzen 3900X in my home server. But coming back to the 8gb of RAM of the new Raspberry Pi Model 4. For my purposes these 8 gb are well suited. So far I haven't maxed them out and I think with only four CPU cores it probably will never happen. However, the overall process of running the analysis in parallel on multiple nodes of a small cluster was more than easy with the help of GNU parallel. For more possibilities and documentation see GNU parallel - Remote execution. As a first test and first excursion in the realm of cluster computing I'm impressed with the simplicity and robustness of this program.
References
[1] O. Tange (2018): GNU Parallel 2018, March 2018, https://doi.org/10.5281/zenodo.1146014
≡