Searching for NLRs
NLRextract - search for NLR related HMMs in protein sequences
NLRextract code and install instructions can be found on the GitHub page: https://github.com/Daniel-Ze/NLRextract
What are NLRs and why search for them in proteoms
NLR stands for "NOD-like Receptors" or "Nuncleotide Binding Leucine Rich Repeats" depending on which publication you stumble upon. These genes form gene families of sometimes several hundred genes per genome in plants. However they are not exclusive to plants. These genes were first described in mammalian cells and were shown to be responsible for immune responses by functioning as receptors that can induce apoptosis of cells. For the NOD-like receptors a so-called apoptosome structure has been described which is a oligomer of NLRs forming a circle with a pore-like structure in the center [1][2]. For plants a similar structure was proposed and could be proven in recent years. The structure was termed resistosome and it was first described for the NLR ZAR1 from Arabidopsis thaliana. After recognition of the effector uridylylated PBL2 protein by ZAR1, the protein switches to its active state and forms together with RKS1 and PBL2 an hetero-oligomer (hetero-pentamer to be precise) like the mammalian apoptosome [3][4]. Furthermore, a recent publication could confirm that the described resistosomes form ion channels that localize in the cell membrane and are permeable for Ca2+ ions. The influx of Ca2+ then leads to cell death [5].
Fig.1 - Differnt modes of actions of NLRs. [6]
ZAR1 represents the guard hypothesis mode of action shown for NLRs. Here the NLR is guarding a plant protein which can be targeted by effectors of the pathogen. Modifications of the guarded protein by the pathogen effector are registered by the NLR, leading to activation and subsequent immune response. This can also happen with protein decoys that mimic the actual effector target. In some scenarios, an additional N-terminal domain of an NLR poses as effector target. A modification of this decoy domain causes the NLR and the subsequent immune response to be reactivated. Knowledge about these functions emerged only in recent years. The best known and best studied mode of action is the direct interaction of effectors with NLRs. However, this is thought to be rarer than initially assumed as it would require a huge number of NLRs to cover the diversity of the fast evolving effectors. The guard hypothesis is thought to be the common mode of action for NLRs. Effectors from different pathogens mostly target similar proteins in plants, which have been termed hubs. It is easier for plants to monitor these hubs than to evolve new immune receptors for each new effector that emerges over time.
Tools to identify NLRs in genomes
Due to their immune response nature, these genes are of immense interest in terms of breeding new resistant cultivars. It is therefore not surprising that the characterization of the "NLRome" has become a standard task for novel genomes. There are several tools out there that do this job.
Tab. 1 - Tools for the detection of NLRs in genomes
| Tool | Genetic data | Reference |
|---|---|---|
| NLR-Parser | Genome, Proteom | https://doi.org/10.1093/bioinformatics/btv005 |
| NLR-Annotator | Genome | https://doi.org/10.1104/pp.19.01273 |
| NLGenomeSweeper | Genome | https://dx.doi.org/10.3390%2Fgenes11030333 |
| NLRtracker | Transcriptome,Proteom | https://doi.org/10.1101/2020.07.08.193961 |
NLR-Parser
NLR-Parser is the most prominent one of the tools out there. Under the hood, NLR-Parser is using custom NLR related motifs generated with the MEME suite. The search is performed with the tool mast from the MEME suite and the custom meme.xml file. The overall performance is fast and no multi-threading is required. The installation is a bit of a pain (at least for the older version of NLR-Parser) as a specific version of the MEME suite is required which is not available on the conda network meaning you have to compile it yourself (if you manage...). However, a new version of NLR-Parser is available that is not dependent anymore on the old MEME suite version. The output you get is a table with the NB-ARC boundaries and the grouping into complete and partial CNLs and TNLs.
NLR-Parser example output
SequenceName class complete start end NB-ARC 2-NBLRR-Signal MotifList
Solyc10g008240.3.1 CNL partial Solyc10g008240.3.1:163 Solyc10g008240.3.1:838 IGPPSKLDVVSIVGMGGIGKTTLARKVYDDIYMEHHFYVRAWITVSQMHQHREMLLGILRCFSLVNDYTYLKSTEQLAEQVYRSLKGRRYLIAMDDVWDTTAWDVVKRSFPDDKNGSRVILTSRLANVGIYASSGSPPHYMRCLSVDRSLKLFNLKVFGRENCPLELEKATKQIVGKCQGLPLAIVVVAGFCSKISKTENCWEDVAHKIGL false 1,6,4,5,10,3,12,2,8,7,9,11
Solyc09g098130.2.1 CNL complete Solyc09g098130.2.1:249 Solyc09g098130.2.1:1245 IRGTNELDVVPIVGMGGQGKTTIARKLYNNDIIVSRFDVRAWCIISQTYNQRELLQDIFSQVTGFNDNGATVDVLADMLRRKLMGKRYLIVLDDMWDCMVWDDLRLSFPDVGIRSRIVVTTRLEEVGKQVKYHTDPYSLPFLTTEESCQLLQKKVFQKEDCPPELQDVSQAVAEKCKGLPLVVVLVAGIIKKRKMEESWWNEVKDALFDYL false 4,17,16,1,6,4,5,10,3,12,2,8,7,9,11
NP_001329653 TNL partial NP_001329653:601 NP_001329653:1365 CIESLDVRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKDLHKEVEVKGHDAVRENFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVNDYRDVDTFLGTLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQIVLSPEVYKTLSLELVKFSNGNPQVLQFLSSIDREWNKLSQEVKTTSPIYIP false 18,13,1,4,5,9,11,11,11,11,9,11,11,9
NLRtracker
A new rising star could be NLRtracker developed by the workgroup of Sophien Kamoun. The installation is straight forward. You need InterProScan (well documented, but a lot of data), a recent version of the MEME suit (conda has you covered!) and R (you'll most probably have that already installed 😉 ). It includes not only the distinction between CNL and TNL NLRs but includes also a subgrouping according to additional domains. This is achieved by generating a comprehensive NLR database of experimentally validated NLRs. From this list they extracted InterProScan domains which they use together with NLR motifs generated with the MEME suite that have been previously published [7]. The overall output is very comprehensive:
(base) daniel@server:~/Programs/NLRtracker$ tree -h NLRtracker_TAIR10/
NLRtracker_TAIR10/
|-- [ 28K] Domains.tsv
|-- [ 59K] NBARC.fasta
|-- [ 48K] NBARC_deduplictated.fasta
|-- [ 55K] NLR-associated.gff3
|-- [ 540] NLR-associated.lst
|-- [214K] NLR.fasta
|-- [887K] NLR.gff3
|-- [2.6K] NLR.lst
|-- [ 14K] NLR_associated.fasta
|-- [ 12K] NLRtracker.tsv
|-- [4.0K] fimo_out
| |-- [ 63M] cisml.xml
| |-- [7.3M] fimo.gff
| |-- [487K] fimo.html
| |-- [3.4M] fimo.tsv
| `-- [5.1K] fimo.xml
`-- [ 86M] interpro_result.gff
Find some more detailed description on the respective GitHub page: NLRtracker
NLRextract
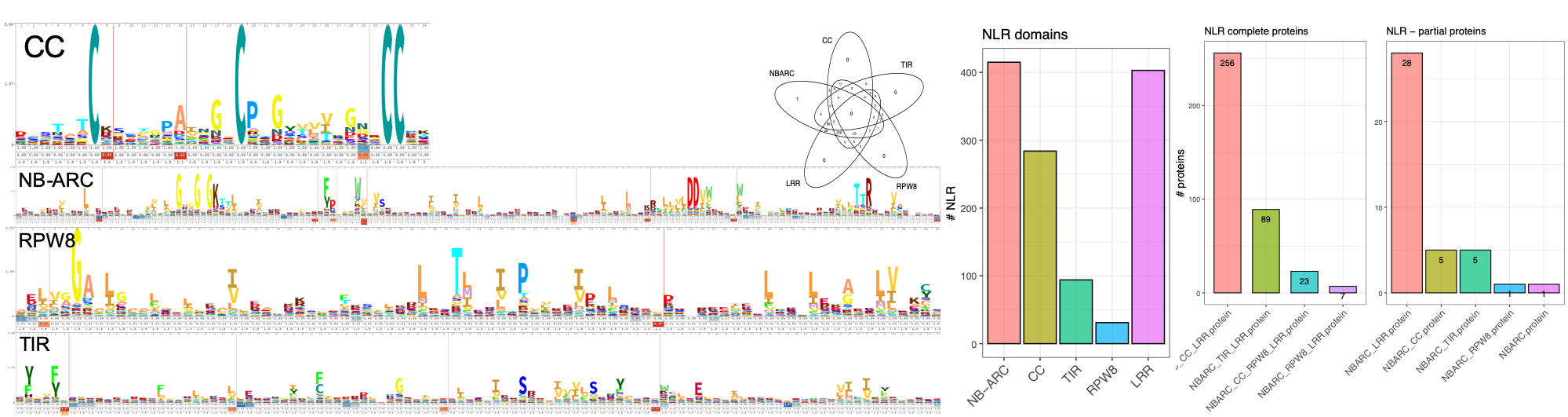
I wrote NLRextract as a quick and dirty tool using curated HMMs of key functional domains of NLRs from the Pfam database. Using hmmer the following domains are searched in the supplied proteins:
- NB-ARC - PF00931.22
- CC - PF04942.14
- RPW8 - PF05659.11
- TIR - PF01582.20
- LRR - PF00560.33 (LRR_1), PF07723.13(LRR_2), PF07725.12(LRR_3), PF12799.7(LRR_4), PF13306.6(LRR_5), PF13516.6(LRR_6), PF13855.6(LRR_8), PF14580.6(LRR_9)
As LRRs are the most variable part and are in no way exclusive for NLRs, several Pfam HMMs were selected that showed association with NLRs. After having identified the different domains in the supplied proteins, a simple Venn diagram is generated and protein sequences are grouped according to their identified domains. As a little plus the MADA domain which was recently identified in so-called helper NLRs of the CNL type (CC domain containing NLRs) is searched [8]. I see the main benefit of using this script in the minimal installation footprint. The only dependencies are R with some libraries (ggplot2, gplots), hmmer (hmmscan and hmmsearch), bedtools (getfasta) and calustalo / clustalw2. Compared to the other tools with InterProScan (5Gb of data to download only for that) or complicated compilation of old software versions (MEME 4.9.1...) it should be quite hassle free to get it running on a standard Ubuntu LTS 20.04 or MacOS Big Sur installation.
What you get when you run the NLRextract:
- plots: the times one of the domains was identified, the complete NLRs, the partial NLRs, everything together
- fasta: the sequences which were identified as NLRs and the functional domains.
- functional domains: Sequences of the identified functional domains are extracted. The extracted domain sequences are aligned and a quick phylogenetic tree is constructed (clustalo and clustalw2).
Comparing the different tools
No new tool gets away without a proper comparison to the available competition. Therefore, I used the TAIR10 protein dataset of the A. thaliana genome and ran NLR-Parser, NLRtracker and NLRextract with it [9]. For an initial comparison I checked the time needed for analyzing the whole proteom of the TAIR10 assembly of A. thaliana. The analysis of all tools was run on a 12 core / 24 thread AMD Ryzen 3900+ and 32 Gb RAM. For NLRtracker and NLRextract 5 cores were chosen in the settings for running the tools.
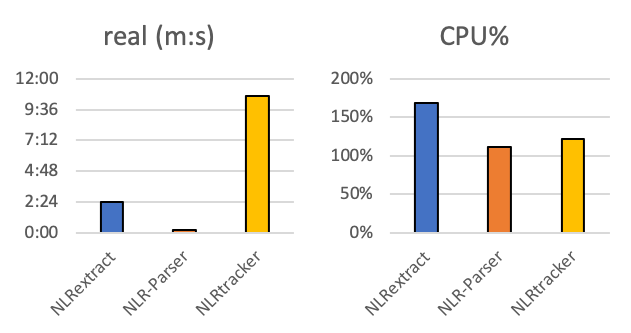
Fig.2 - Time required to complete the A. thaliana proteom analysis and the CPU usage while running the programs.
But what everybody is asking now: Daniel that's nice and all but how are the real results looking?! Well the total amount of identified NLRs is similar for all three tools. Roughly 270 protein sequences were classified as NLRs (including isoforms of genes). NLRextract identfied the smallest number and NLRtracker the highest number of NLR sequences (Fig. 3A). The obtained lists of identfied NLRs were intersected with each other using a Venn diagram (Venny 2.1). All three tools had 247 sequences in common. NLRtracker and NLRextract shared 13 sequences from 12 genes which were not identified by NLR-Parser. On the other side NLRtracker and NLR-Parser shared four sequences. All tools had unique sequences that were not identfied by the others. NLR-Parser showed the highest number with 17 unique sequences, followed by NLRtracker (7) and then NLRextract (2) (Fig. 3B).
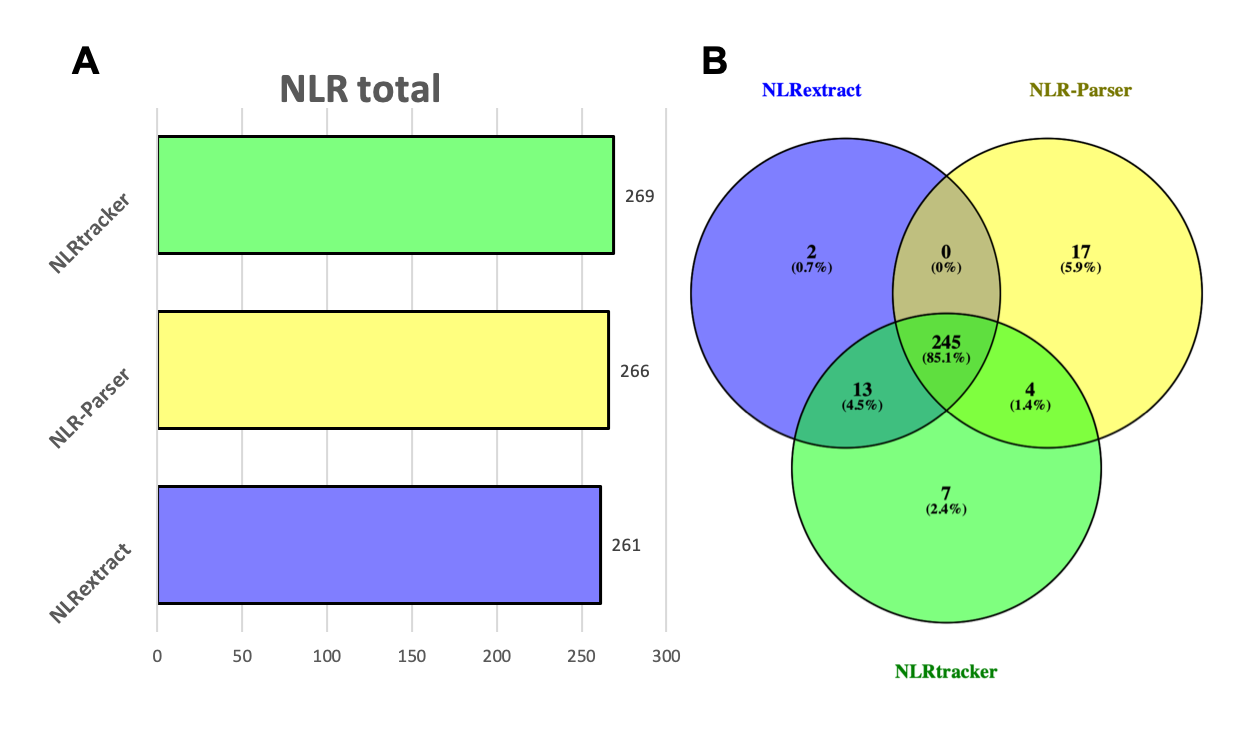
Fig.3 - A Total number of protein sequences identfied as NLRs by the three tools. B Venn diagram of the identfied NLR sequences of the three tools.
To quickly check the functional domain architecture of the shared and unique sequences, they were submitted to conserved domain search (CD-Batch Search). The majority of sequences shared between NLRextract and NLRtracker were related to RPW8 domain containing NLRs (10 out of 13) which are not searched for by NLR-Parser. The other three sequences were partial TIR domain containing NLRs (Sup.Tab. 1). The four sequences shared by NLRtracker and NLR-Parser were LRR containing protein sequences with no other NLR related functional domain annotated (Sup.Tab. 2). One of the two unique sequences of NLRextract shows an annotated CC domain with a low e-value which was probably below the cut-off from NLRtracker and NLR-Parser. For the second sequence a NB-ARC domain was annotated, although with a very low e-value also in this case. The seven unique sequences of NLRtracker are either proteins with annotated LRR domain or with a domain of unknown function (DUF1221) but no further NLR related functional domains (Sup.Tab. 3). NLR-Parser showed the most unique sequences when compared to NLRtracker and NLRextract (Fig. 3B, 17). All except two sequences showed annotated LRR domains without any further NLR related functional domain. The two sequences without LRR showed no further NLR related domains but ATPase activity which could indicate a small cut-off problem for the NB-ARC domain as it was seen for NLRextract (Sup.Tab. 4).
What's the benefit of NLRextract?
So what's the benefit of using NLRextract you might ask yourself. Next to the NLR identfication, NLRextract also performs sequence alignment of the NLR associated functional domains and generates phylogenetic trees from the alignments. This is useful if you want to run these kinds of analyses in a standardized way for a large set of proteoms. Additionally the MADA motif is identified in CNL NLRs which is also missed by NLR-Parser (of course this is included in NLRtracker 😉).

Fig.3 - Phylogenetic trees of the NLR functional domains. The trees were visualized with iTOL.
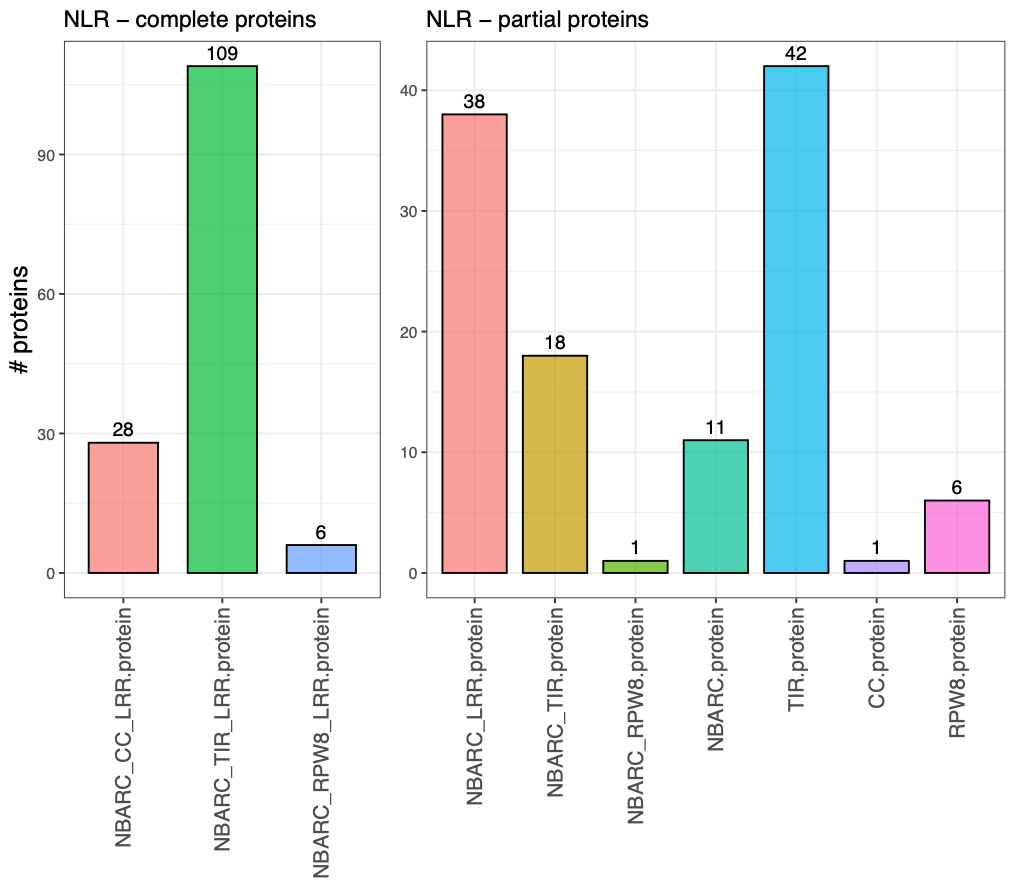
Fig.4 - Number of complete and partial NLRs identfied by NLRextract.
Conclusion
Overall NLRextract performs very well compared to the two other tools. It identfies all the sequences NLR-Parser does plus it identifies the RPW8 domain containing NLR protein sequences that are identfied by NLRtracker. One of the two unique protein sequences is a truncated protein with a annotated CC domain missing in the annotation of NLRtracker and NLR-Parser. Only one protein was misidentfied. NLRextract runs faster than the more comprehensive NLRtracker. NLRtracker is designed to detect novel domains in between the classical NLR functional domains which is an important step for novel findings. However, running it on a large set of proteoms can cause problems in large scale analyses. A preliminary version of NLRextract is freely available as a GitHub repository NLRextract. All the described results from this article will be included in a updated version of the repository in a short while. If you have ideas or concerns about the tool write me an email or contact me on the irc server in the channel #bioinf!.
References
[1] Hua Zou, Yuchen Li, Xuesong Liu, Xiaodong Wang, 1999, An APAF-1 Cytochrome-c Multimeric Complex Is a Functional Apoptosome That Activates Procaspase-9*. Journal of Biological Metabolism, Vol. 274, Issue 17, P11549-11556. https://doi.org/10.1074/jbc.274.17.11549
[2] Shujun Yuan, Xinchao Yu, Maya Topf, Steven J. Ludtke, Xiaodong Wang, Christopher W. Akey, 2010, Structure of an Apoptosome-Procaspase-9 CARD Complex. Structure, Vol 18, Issue 5, P571-583. https://doi.org/10.1016/j.str.2010.04.001
[3] Hayden Burdett, Adam R. Bentham, Simon J. Williams, Peter N. Dodds, Peter A. Anderson, Mark J. Banfield, Bostjan Kobe, 2019, The Plant "Resistosome": Structural Insights into Immune Signaling. Cell Host & Microbe, Vol. 26, Issue 2, P193-201. https://doi.org/10.1016/j.chom.2019.07.020
[4] Jizong Wang, Meijuan Hu, Jia Wang, Jinfeng Qi, Zhifu Han, Guoxun Wang, Yijun Qi, Hong-Wei Wang, Jian-Min Zhou, Jijie Chai, 2019, Reconstitution and structure of a plant NLR resistosome conferring immunity. Science, Vol. 364, Issue 6435. https://doi.org/10.1126/science.aav5870
[5] Pierre Jacob, Nak Hyun Kim, Feihua Wu, Farid El-Kasmi, Yuan Chi, William G. Walton, Oliver J. Furzer, Adam D. Lietzan, Sruthi Sunil, Korina Kempthorn, Matthew R. Redinbo, Zhen-Ming Pei, Li Wan, Jeffery L. Dangl, 2021, Plant "helper" immune receptors are Ca2+-permeable nonselective cation channels. Science, Vol 373, Issue 6553. https://doi.org/10.1126/science.abg7917
[6] Stella Cesari, 2017, Multiple strategies for pathogen perception by plant immune receptors. New Phytologist, Vol. 219, Issue 1, P17-24. https://doi.org/10.1111/nph.14877
[7] Florian Jupe, Leighton Pritchard, Graham J Etherington, Katrin MacKenzie, Peter JA Cock, Frank Wright, Sanjeev Kumar Sharma, Dan Bolser, Glenn J Bryan, Jonathan DG Jones & Ingo Hein, 2012, Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics, 13, 75. https://doi.org/10.1186/1471-2164-13-75
[8] Hiroaki Adachi, Mauricio P Contreras, Adeline Harant, Chih-hang Wu, Lida Derevnina, Toshiyuki Sakai, Cian Duggan, Eleonora Moratto, Tolga O Bozkurt, Abbas Maqbool, Joe Win, Sophien Kamoun, 2019, An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife, 8:e49956 http://dx.doi.org/10.7554/eLife.49956
[9] Arabidopsis thaliana version TAIR10 proteom https://www.arabidopsis.org/download_files/Proteins/TAIR10_protein_lists/TAIR10_pep_20101214
